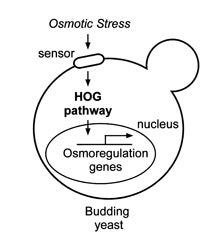
 A fundamental problem in biology is how cells sense and respond to stimuli such as changes in pressure, osmolarity, or mechanical forces. Most of our research employs the yeast Saccharomyces cerevisiae as a model system for understanding the molecular mechanisms required for sensing and responding to changes in external osmolarity and other stresses. Genetic and biochemical analysis of mutants that fail to grow in high (or low) osmolarity media has allowed us to uncover two stress-activated signaling pathways, the hog and cell intensity pathways, respectively. Both of these pathways contain a unique map kinase cascade. We are currently investigating the mechanism of osmosensing and transcriptional regulation by these pathways plus their role in osmotic stress-induced cell cycle delay. A pathway similar to the yeast hog pathway is found in osteoblasts, mammalian cells not usually exposed to osmotic stress. We are therefore investigating the role of the hog pathway in responses of these cells to mechanical stress. In addition, my research group, in collaboration with Dr. Mikos (Bioengineering, Rice U.), is examining the function of the mammalian hog pathway in mediating activation of bone forming cells by mechanical stress.
A fundamental problem in biology is how cells sense and respond to stimuli such as changes in pressure, osmolarity, or mechanical forces. Most of our research employs the yeast Saccharomyces cerevisiae as a model system for understanding the molecular mechanisms required for sensing and responding to changes in external osmolarity and other stresses. Genetic and biochemical analysis of mutants that fail to grow in high (or low) osmolarity media has allowed us to uncover two stress-activated signaling pathways, the hog and cell intensity pathways, respectively. Both of these pathways contain a unique map kinase cascade. We are currently investigating the mechanism of osmosensing and transcriptional regulation by these pathways plus their role in osmotic stress-induced cell cycle delay. A pathway similar to the yeast hog pathway is found in osteoblasts, mammalian cells not usually exposed to osmotic stress. We are therefore investigating the role of the hog pathway in responses of these cells to mechanical stress. In addition, my research group, in collaboration with Dr. Mikos (Bioengineering, Rice U.), is examining the function of the mammalian hog pathway in mediating activation of bone forming cells by mechanical stress.

WEBSITE(S)| PubMed Publication List
Research Areas
Molecular genetics and biochemistry of signal transduction
Education
B.A. Biology (1974) Johns Hopkins University
Ph.D. Cell Biology (1981) Yale University
