The goal of the Chappell lab is to forward our ability to understand and engineer the bacteria domain of life. Central to this is our ability to control how cells express their genetic code. Our lab focuses on understanding how the biomolecule RNA can be designed to create synthetic regulators of gene expression—allowing for the manipulation of natural cellular processes to elicit deeper biological understanding and for the engineering of new synthetic cellular functions. As such our lab focuses both on the creation of new gene regulatory tools and their application.
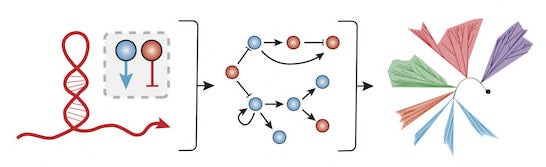
Central to our approach is the philosophy that RNA is a powerful molecule for cellular engineering. Firstly, RNA regulators are highly designable, as they exert regulation by the formation of specific structures within mRNAs that are determined largely by the simple rules of Watson-Crick base pairing. Secondly, RNA regulators can be used to create gene regulatory networks capable of performing complex signal processing. Such synthetic networks are highly valuable to many Synthetic Biology applications, such as metabolic engineering and creation of biological diagnostics. Finally, RNA regulators are potentially highly transferable across bacterial species because they depend on universal base pairing interactions between nucleotides and require minimal cellular machinery. Therefore, RNA is potentially a powerful molecule for harnessing the diversity of the bacteria domain of life.
The main areas of research focus are currently: (1) Creation of synthetic RNA regulators of gene expression. (2) Deciphering the portability of RNA regulators across the bacteria domain of life. (3) Creation of synthetic genetic circuits capable of performing signal processing. (4) Applying RNA-based tools for functional genomics

